Skyline supports several workflows where the retention time of peptide search identified MS/MS spectra are used to help it pick chromatogram peaks, and for subsequent visual inspection. The most visible effect of when this extra information is present and usable by Skyline is the addition of ID annotations to the chromatogram graphs, as shown below:

If you build a spectral library or use the Import Peptide Search wizard to import Mascot search results for use with full-scan chromatogram extraction, and find that you do not see ID annotations in your chromatograms as you would expect, the problem most likely originates with the MGF converter you used to create MGF files as input to Mascot.
For Skyline to be able to place an identified spectrum on an extracted chromatogram, it needs two things, beyond the peptide identification itself:
The spectrum source file name does not need to match exactly with the file specified in the Import Results form. Skyline uses base name matching, which counts all of the following files as matching:
Note also that Skyline completely ignores any path information included with the spectrum source file name. Many converters will include a full path, but this is not necessary, and Skyline will match chromatogram data imported from any path, as long as the file basenames match.
The first place to look for clues on contents of any library is the Skyline Spectral Library Explorer (View > Spectral Libraries). A library built from search results that contain the necessary information will look like this:
If you click the button beside the Library drop down list, Skyline will display the Library Details form with a list of the spectrum source files from which there are identified spectra in the library:

The most common issue you will see with a Mascot DAT file is that it does not contain spectrum source file information in a format that Skyline can understand. That format can be traced back to the TITLE lines in your original MGF file. Thanks to a lack of standardization in this area, a long stream of bug reports has lead to Skyline handling a number of different TITLE line formats, but the most flexible and robust format are:
TITLE=...File: "path/to/file.raw"...
or slightly less robust:
TITLE=...File: path/to/file.raw ...
TITLE=...[path/to/file.raw]
Since the first does not allow spaces in the path, and the second does not allow brackets in the path. Note again that the path information will be ignored by Skyline in matching with imported chromatogram files, though characters in the path can have a negative impact on parsing of some formats (e.g. spaces in format that relies on a space as a terminal character).
The retention times are provided by RTINSECONDS lines in the MGF like:
RTINSECONDS=3006.0281
Problems in either of these can cause issues that show up like the following Spectral Library Explorer figures:
Issue 1: The TITLE line in the MGF file did not contain a recognizable format (described above) from which the spectrum source file can be parsed, causing the DAT file name to be used instead. If your DAT file contains the search results for a single file, this can be corrected by simply renaming the DAT file to have the same base name as the data file you will import for chromatogram extraction (e.g. spectrum_source.dat).
Issue 2: The RTINSECONDS line in the MGF file was missing, causing the spectrum RT value to be set to zero.
Issue 3: A time outside the gradient length is shown. Something has gone wrong with the library builder parsing this file. You should report something like this to the Skyline team.

Issue 4: Every spectrum has a different source file not representative of files on disk. Something has gone wrong with the library builder parsing this file. You should report something like this to the Skyline team.
If you run into any problems like this, we always recommend installing ProteoWizard and using MSConvertGUI to create your MGF files, as shown below:
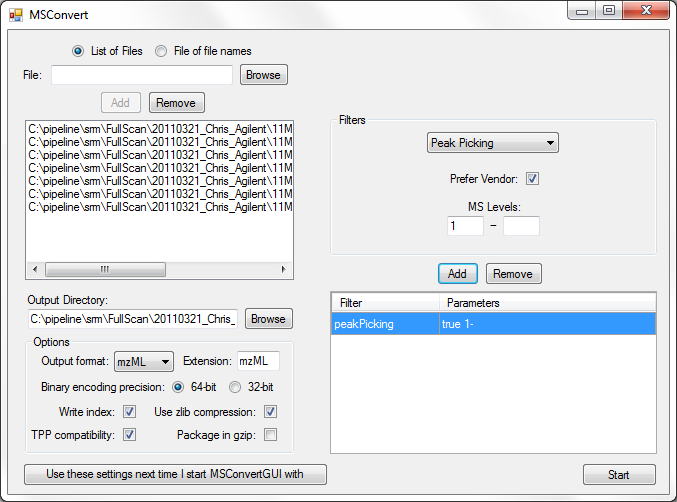
Note that you must make sure the TPP compatibility check box is checked.
If the MGF converter you used comes from an instrument vendor or professional software company, and you want help communicating with them what is required for full integration with a workflow that includes Skyline, either point them to this page, or post your issue to the Skyline support board.