(This tip relates to Skyline 4.2 and later.)
This is a quick demonstration on how to use MSConvert to generate a demultiplexed dataset from an input datafile(s) containing spectra with overlapping data independent acquisition windows. In the case of the overlapped window approach described in this manuscript, the output from MSConvert will contain twice as many spectra as the input (two demultiplexed spectra are generated from each acquired MS/MS spectrum). This tutorial uses MSConvert distributed with ProteoWizard version 3.0.18328 with vendor libraries downloadable here: http://proteowizard.sourceforge.net/download.html
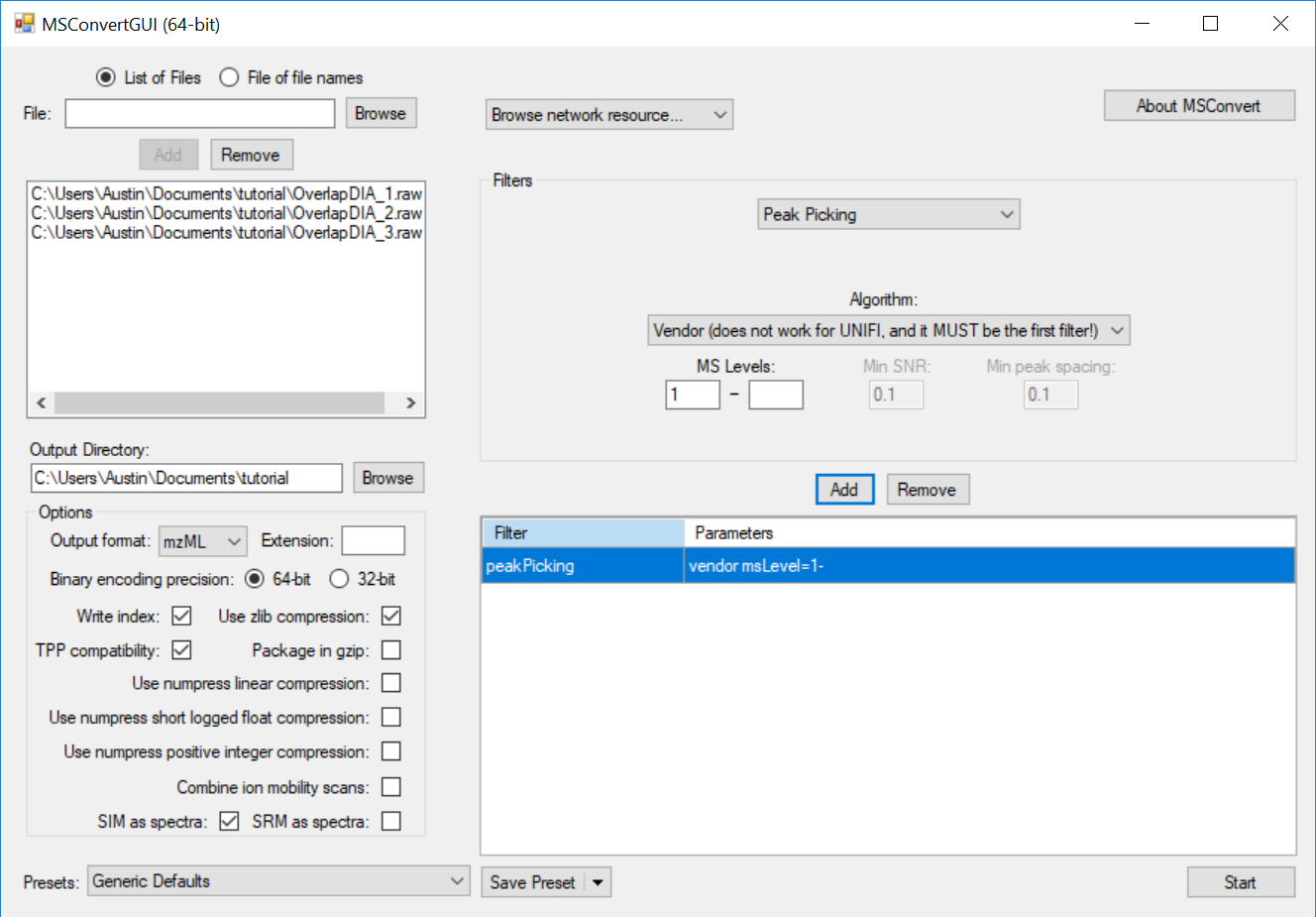
This causes the MS2 data to be centroided prior to demultiplexing, which is currently a requirement for full-spectrum demultiplexing using MSConvert. If the data were acquired with centroiding enabled, this step will have no effect and demultiplexing will proceed as expected.
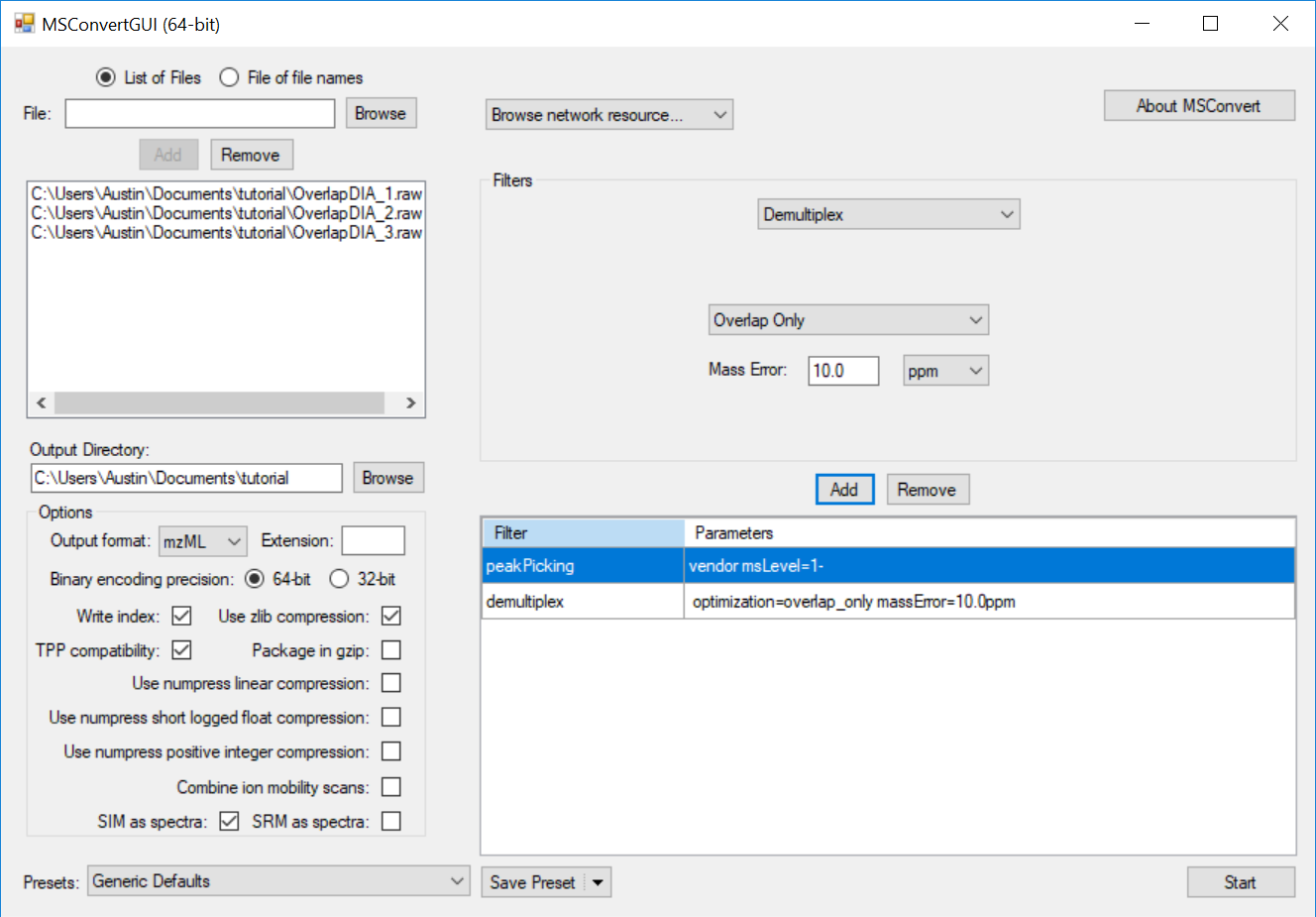
Note that the mass error may need to be adjusted depending on instrument platform. The mass error should be set to the maximum error expected in m/z measurement of the same analyte in subsequent spectra. Note that this measurement is of expected deviation of a measurement from spectrum to spectrum, not its deviation from the correct theoretical m/z (mass accuracy).
| Attached Files | ||