|
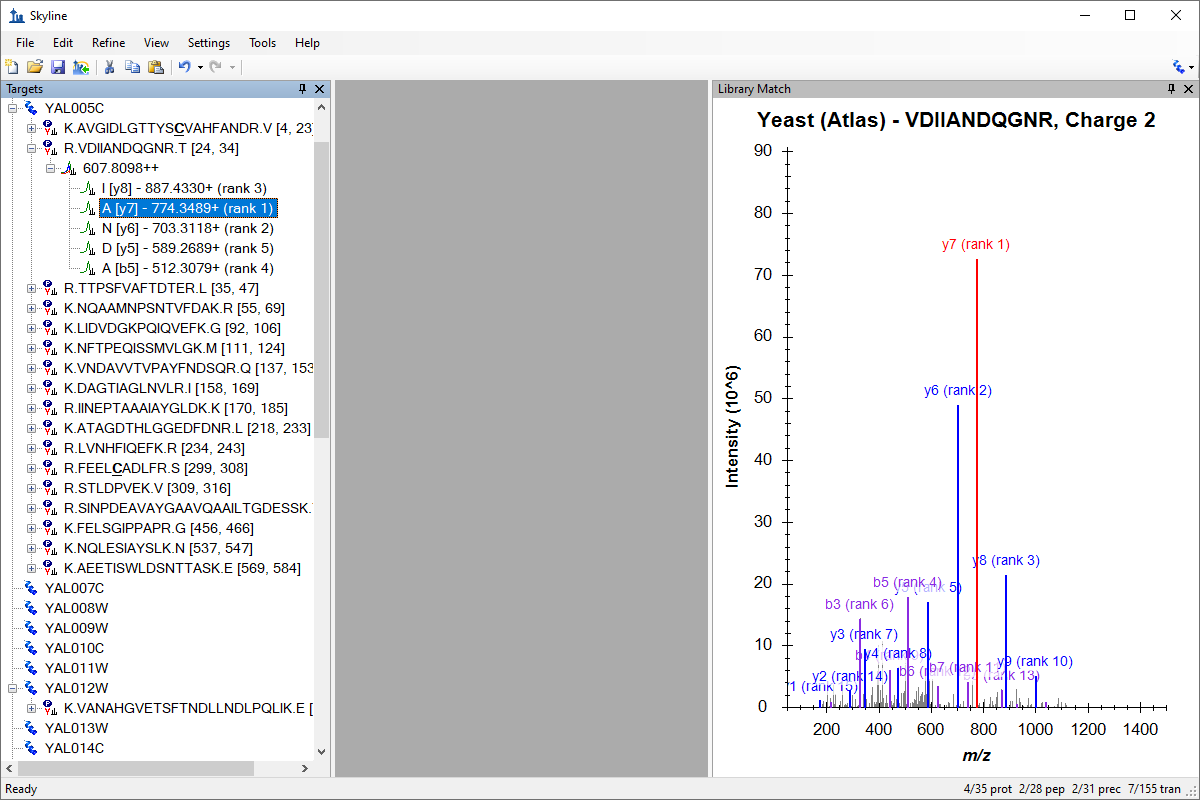
Tutorial: Targeted Method Editing Create a Skyline document for a targeted proteomics experiment from a MS/MS spectral library built with pepXML and mzXML and a background proteome from a FASTA format file. Combine these with a public MS/MS spectral library from the Global Proteome Machine to guide creation of a new Skyline document targeting selected yeast proteins, peptides and product ions. From this document, export a transition list, ready to run on a SCIEX 4000 Q Trap. (26 pages)
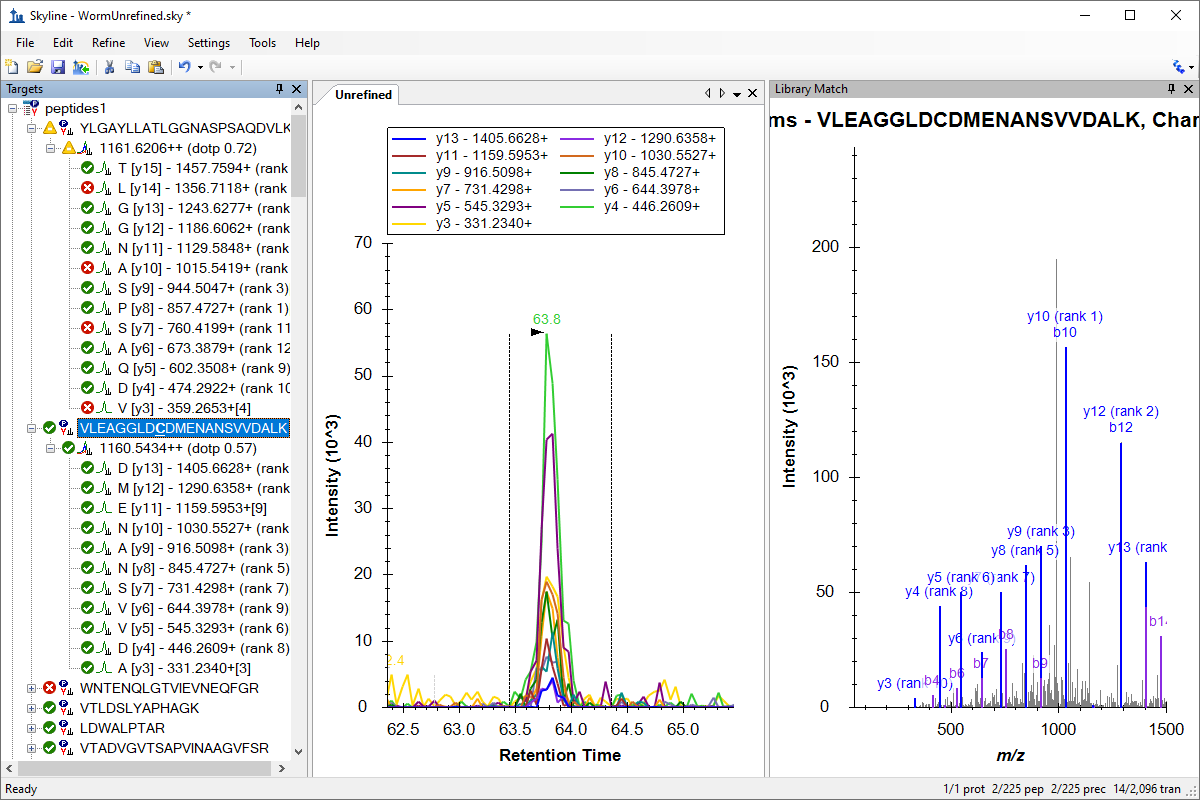
Tutorial: Targeted Method Refinement Refine a broad set of SRM measurements, importing instrument data, and refining the document. Start with an unrefined document requiring over 2000 transitions and 39 injections to measure unscheduled on a Thermo TSQ. Learn how to import all 39 injections into a single replicate. Use the Skyline refinement dialog to remove all but the best transitions for the highest confidence peaks. (28 pages)
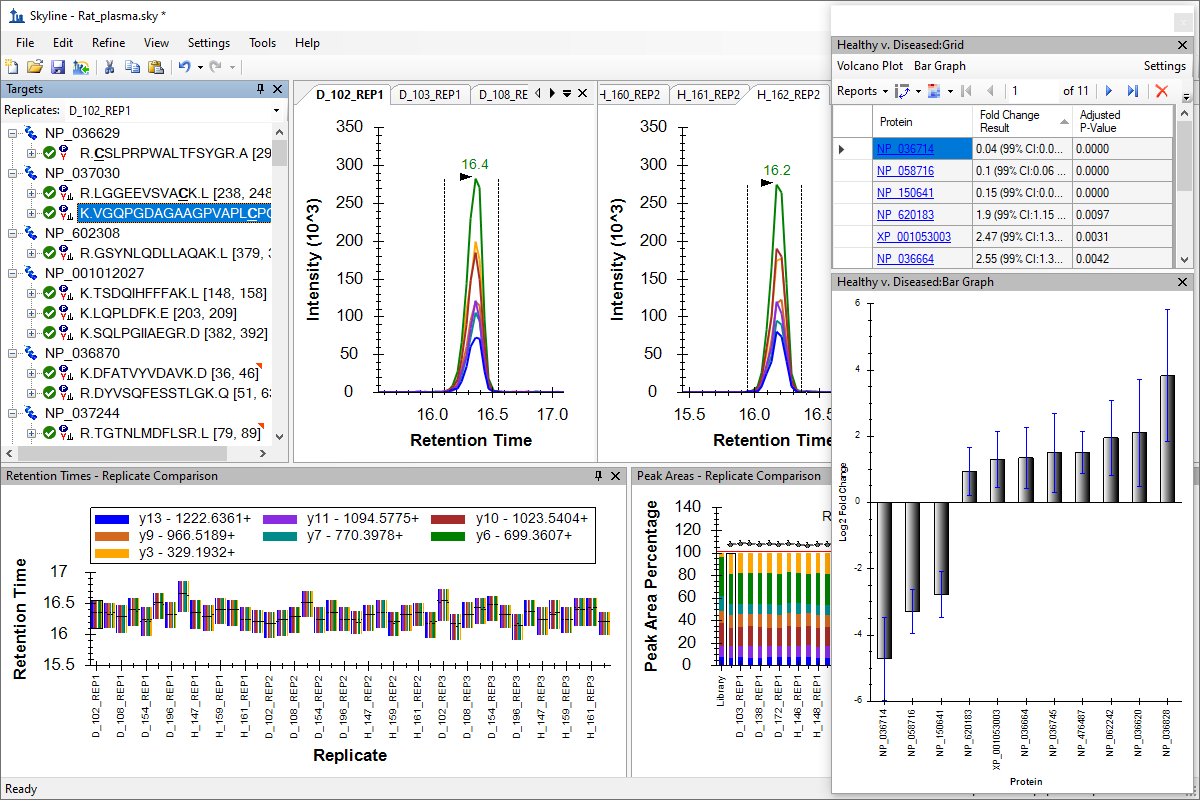
Tutorial: Grouped Study Data Processing Process multi-replicate study data effectively with Skyline. Take an initial set of targets already refined as detectable and further refine this set for evidence of differential abundance between healthy and diseased subjects a study of plasma from 14 rats. Learn to use the powerful Skyline interactive displays to quickly investigate and understand data anomalies. Gain experience with the Skyline Group Comparison framework. (70 pages)
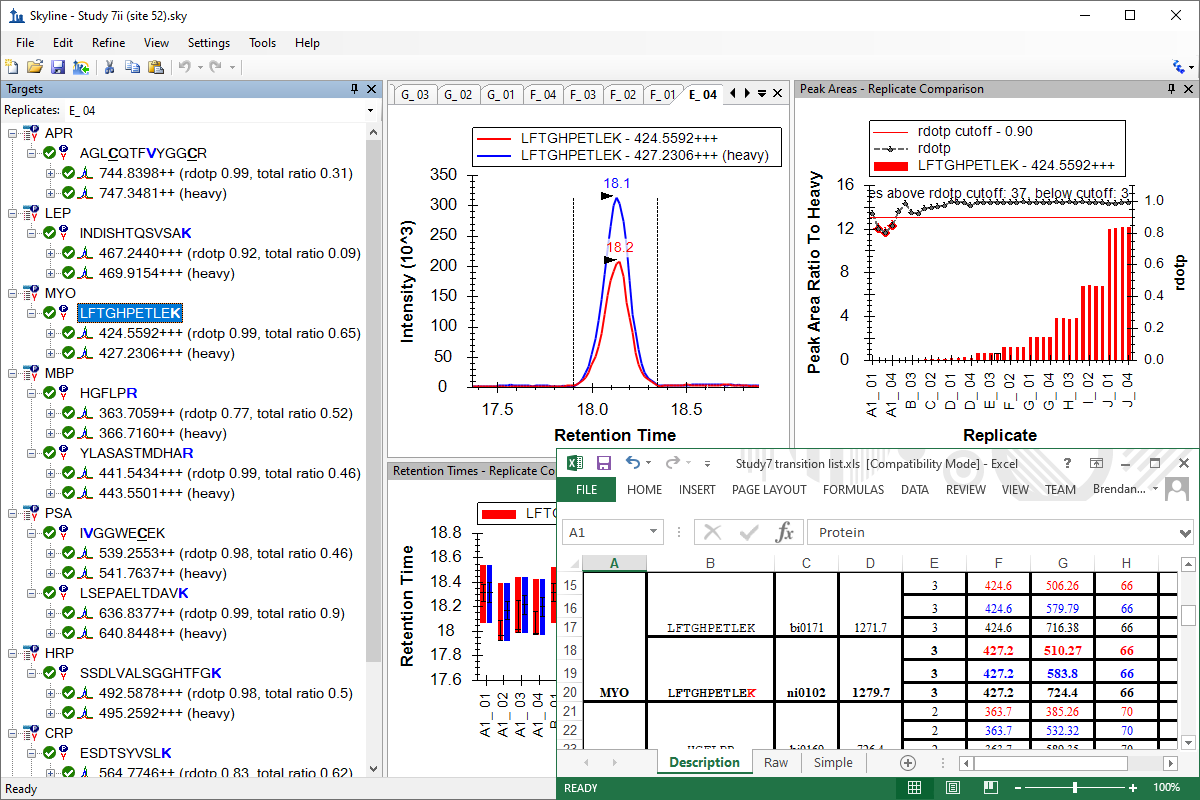
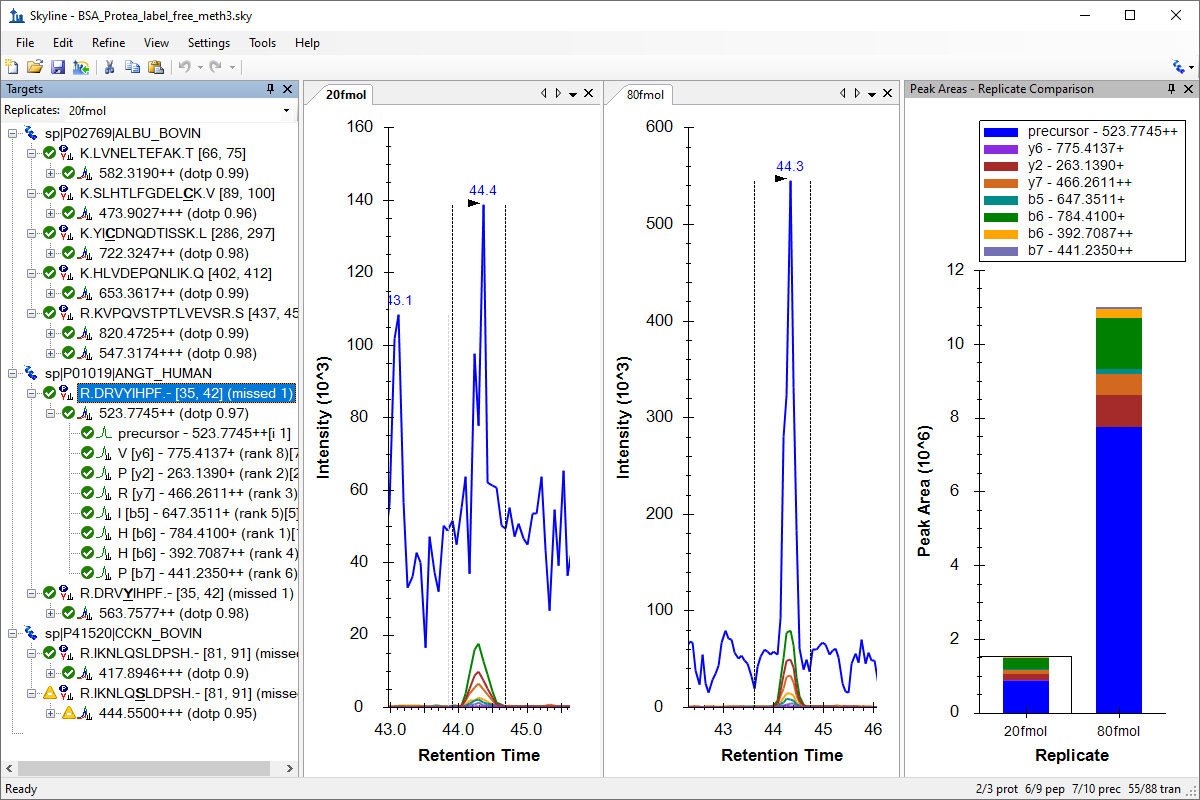
Tutorial: Existing & Quantitative Experiments Work with quantitative experiments and isotope labeled reference peptides, by starting with experiments with published transition lists and SRM mass spectrometer data. Learn effective ways of analyzing your data in Skyline using several of the available peak area and retention time summary charts. (43 pages)
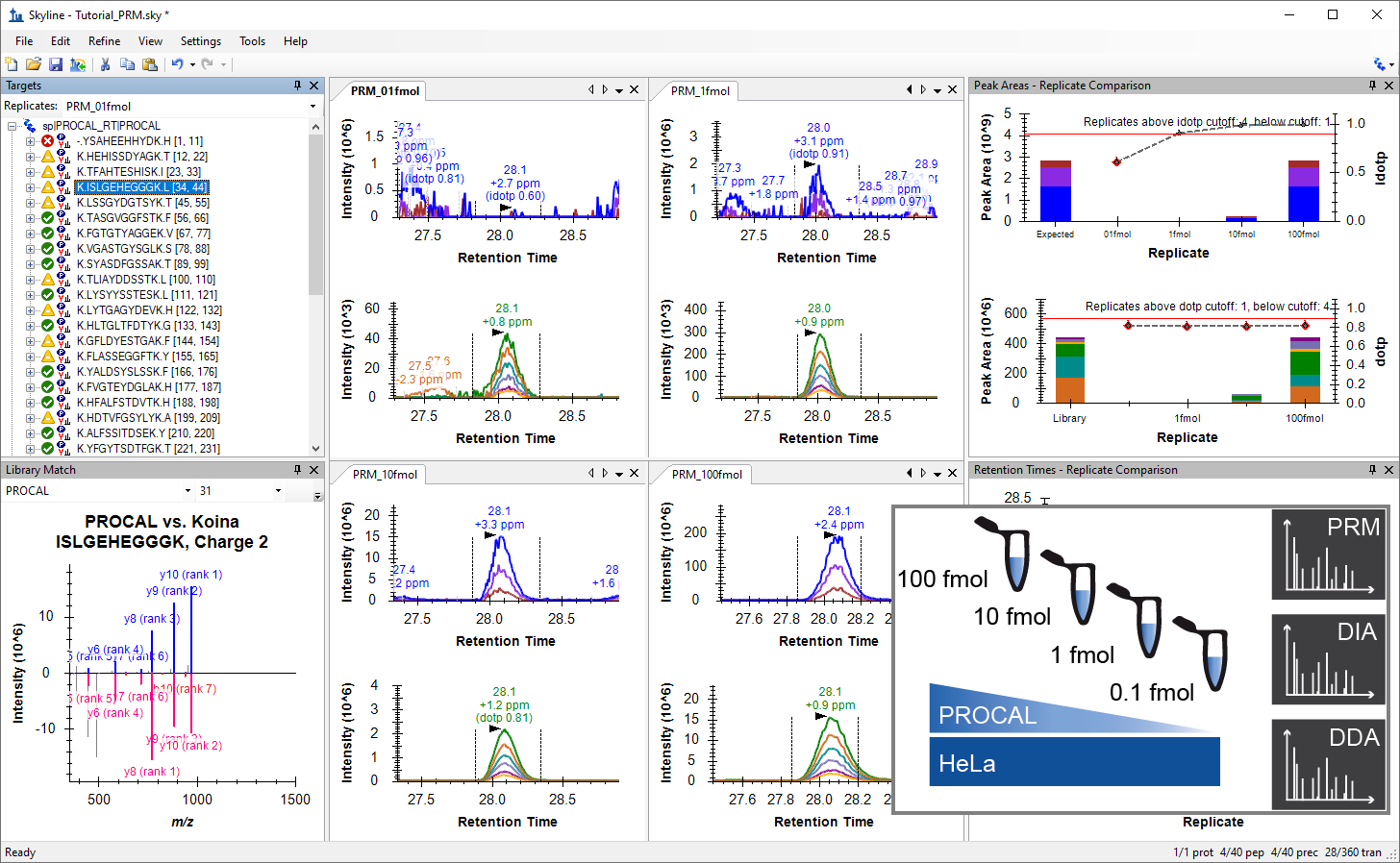
Tutorial: Comparing PRM, DIA, and DDA erform a targeted analysis in Skyline with PRM, DIA and DDA measurement data. The four samples used for this tutorial were generated by mixing HeLa protein digest with PROCAL peptide standard at defined concentrations. Each sample was measured with three technical replicate injections in PRM, DIA and DDA mode on a Quadrupole-Orbitrap-Iontrap (Tribrid) mass spectrometer (Eclipse, Thermo). (38 pages)
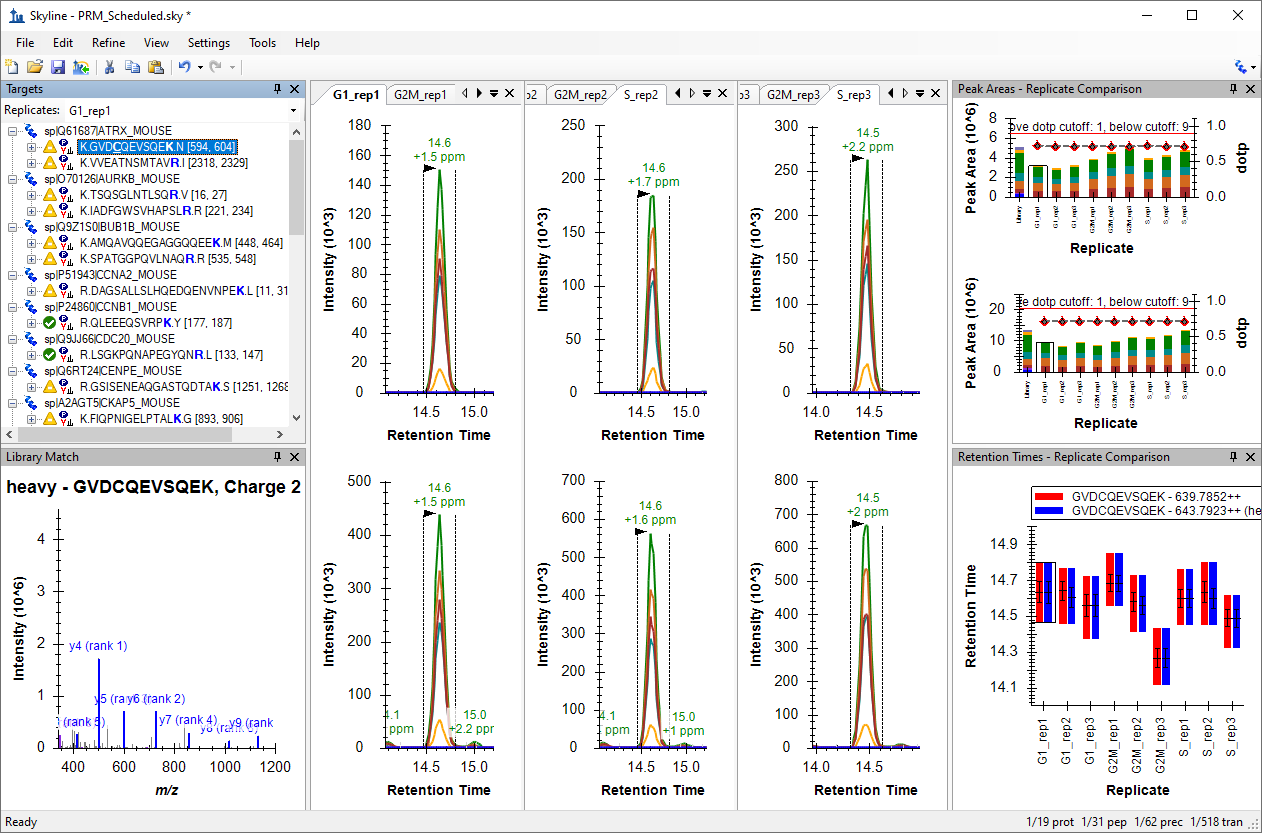
Tutorial: PRM With an Orbitrap Mass Spec Get hands-on experience setting up an acquisition method to quantify 31 peptides from 19 proteins of interest in murine fibroblasts using Parallel Reaction Monitoring (PRM). Work with three biological replicates measured in 9 runs on a Thermo Fusion mass spectrometer employing the Orbitrap mass analyzer. (44 pages)
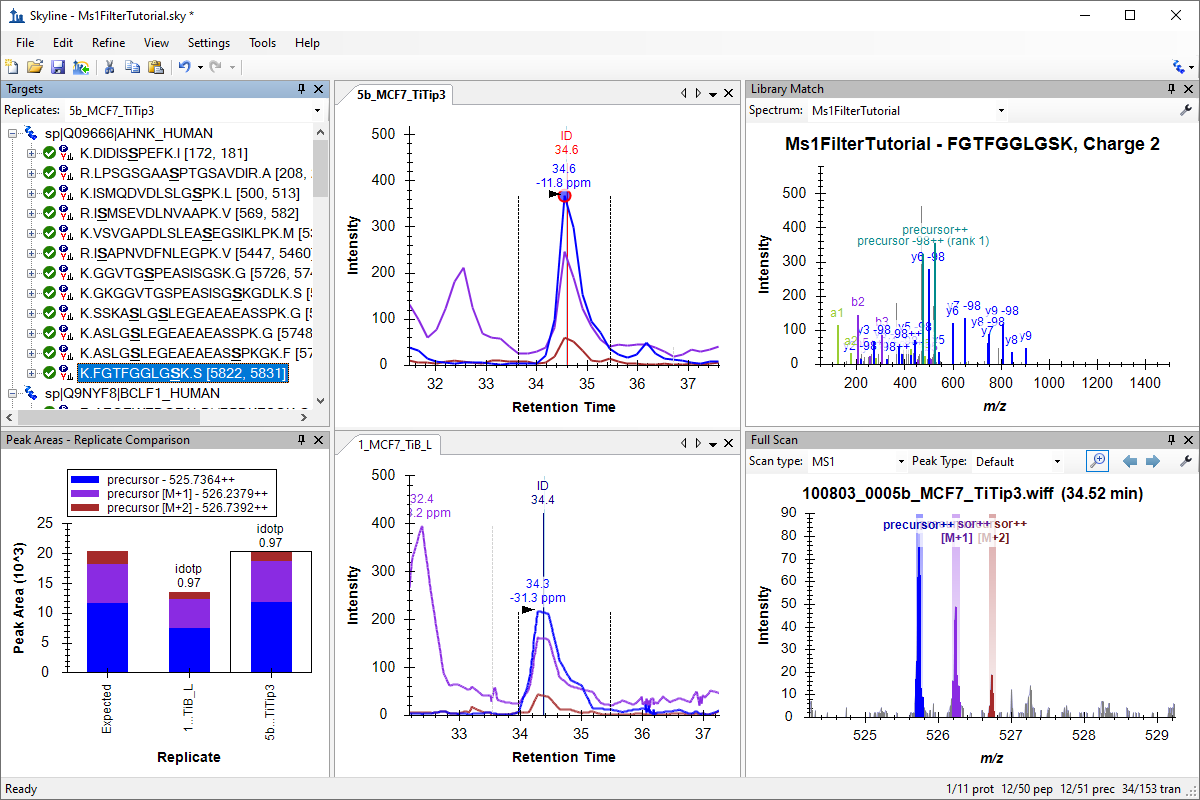
Tutorial: MS1 Full-Scan Filtering Measure quantitative differences in peptide expression using the MS1 scans from your data dependent acquisition (DDA) experiments. Generate a spectral library from a discovery data set, set up a Skyline document for MS1 filtering, import raw mass spectrometer data to extract precursor ion chromatograms from MS1 scans, with peak picking guided by MS/MS peptide identifications, and further process the resulting quantitative data in Skyline. (38 pages)
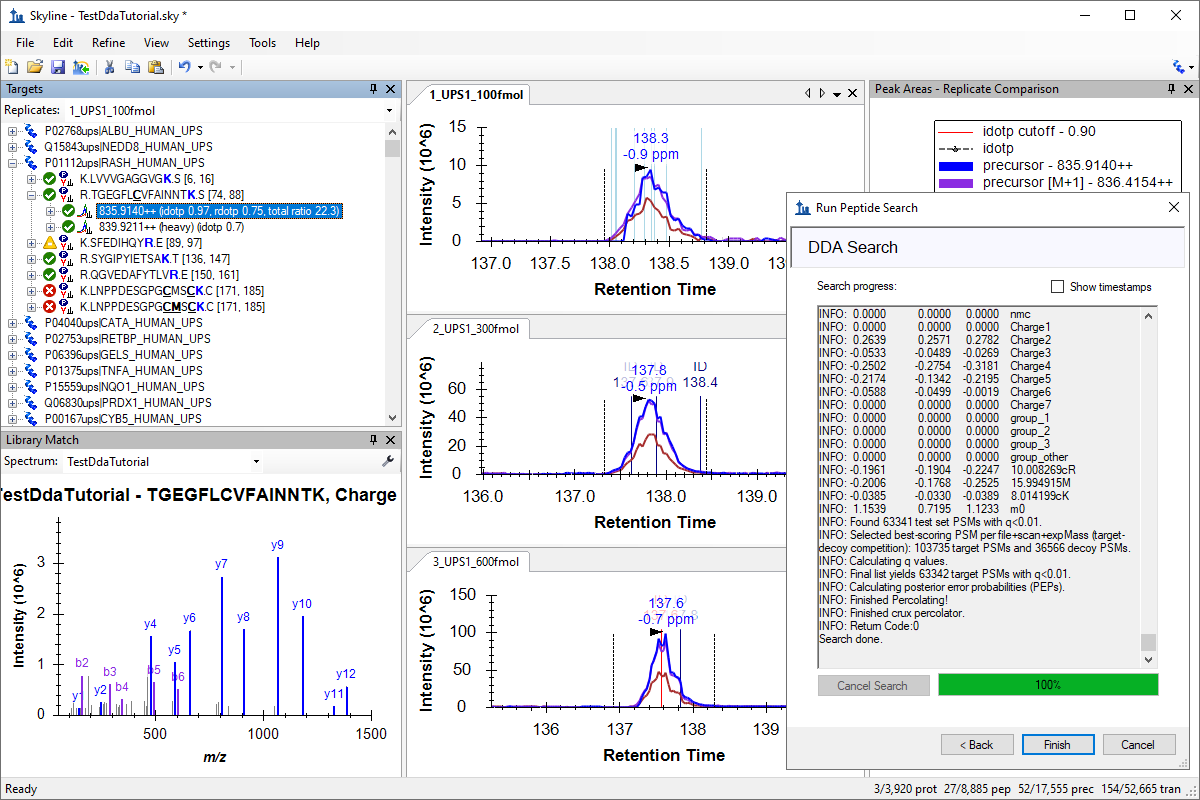
Tutorial: DDA Search for MS1 Filtering Start with DDA mass spectometer files, run a peptide search with MS Amanda, building a spectral library from the search results and finally extract chromatograms for quantitative analysis from the MS1 spectra in the DDA files. (19 pages)
Tutorial: Parallel Reaction Monitoring (PRM) Work with targeted MS/MS (PRM) data acquired on a low resolution Thermo LTQ and a high resolution Agilent 6520 Q-TOF. Gain new understanding of the selectivity and sensitivity differences between peptide quantification using precursor and fragment ions measured on low and high resolution instruments. Discover new ways to work with the rich feature set provided by Skyline for analyzing chromatography-based quantitative proteomics data. (40 pages)
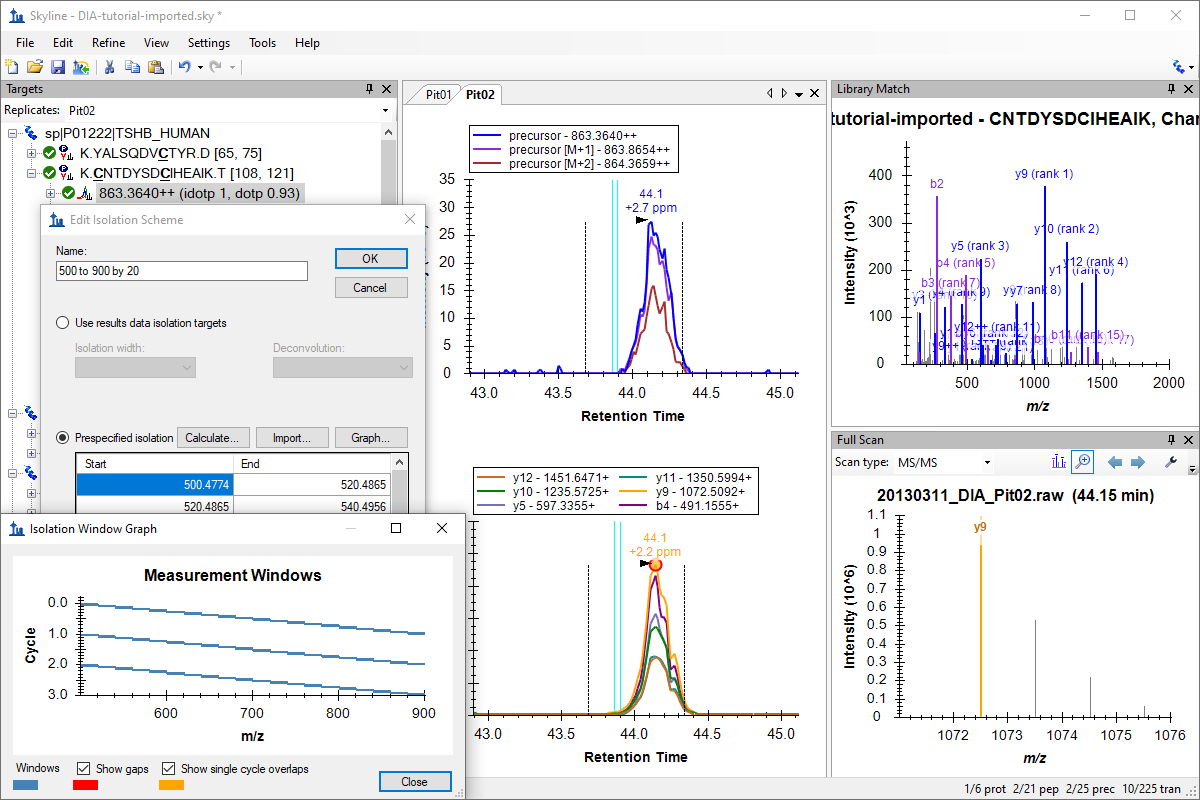
Tutorial: Basic Data Independent Acquisition Work with data independent acquisition (DIA) data, using a workflow that utilizes DIA and DDA runs acquired on the same instrument in series. Define and export a DIA isolation scheme. Build a spectral library from DDA data acquired before the DIA runs for the experiment. Choose peptides and transitions for a target set of proteins based on the spectral library. Import and analyze related DIA runs in Skyline to learn a simple starting workflow for beginning to work with DIA. (40 pages)
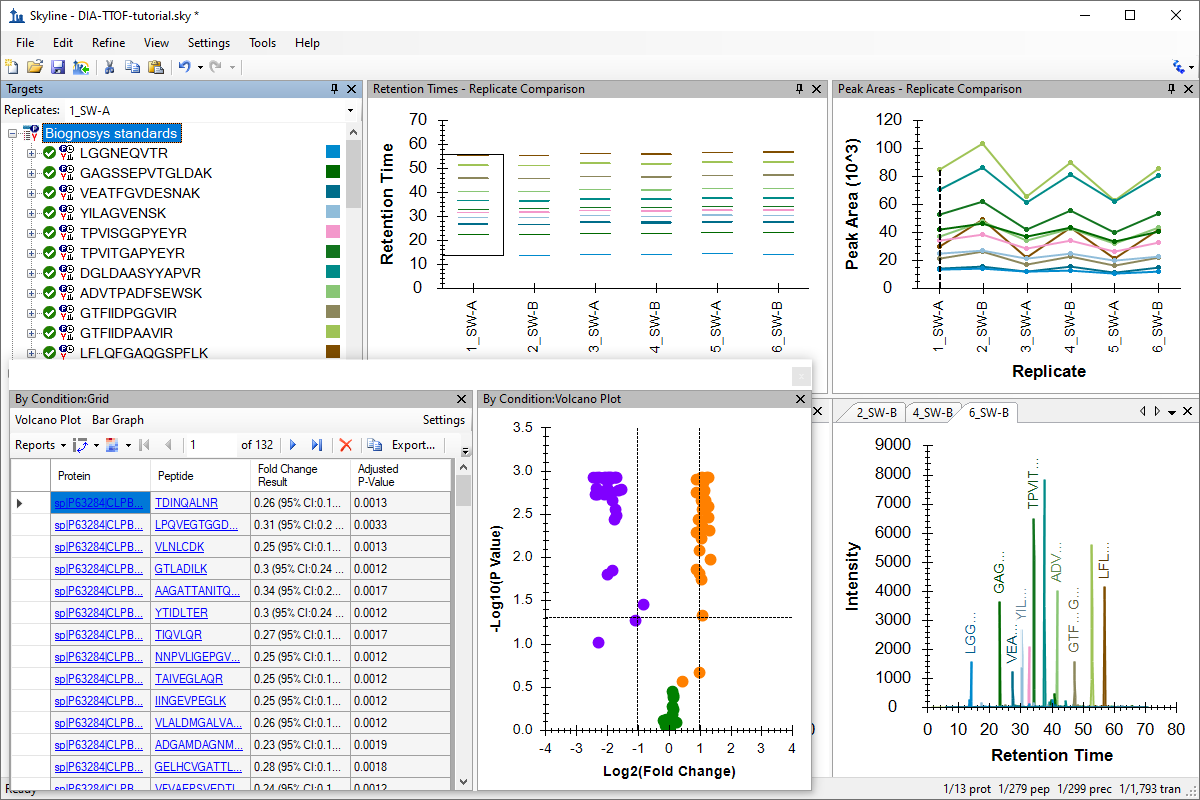
Tutorial: Analysis of DIA/SWATH Data Work with data independent acquisition (DIA) data, from either a Q Exactive or a TripleTOF instrument, using a 3-organism mix data set created for instruction, based on the Navarro, Nature Biotech 2016 benchmarking paper. Use the Import Peptide Search wizard for DIA to build a spectral library from DDA data with automatic iRT calibration for retention time calibration and an mProphet learned model for peptide peak detection. (32 pages)
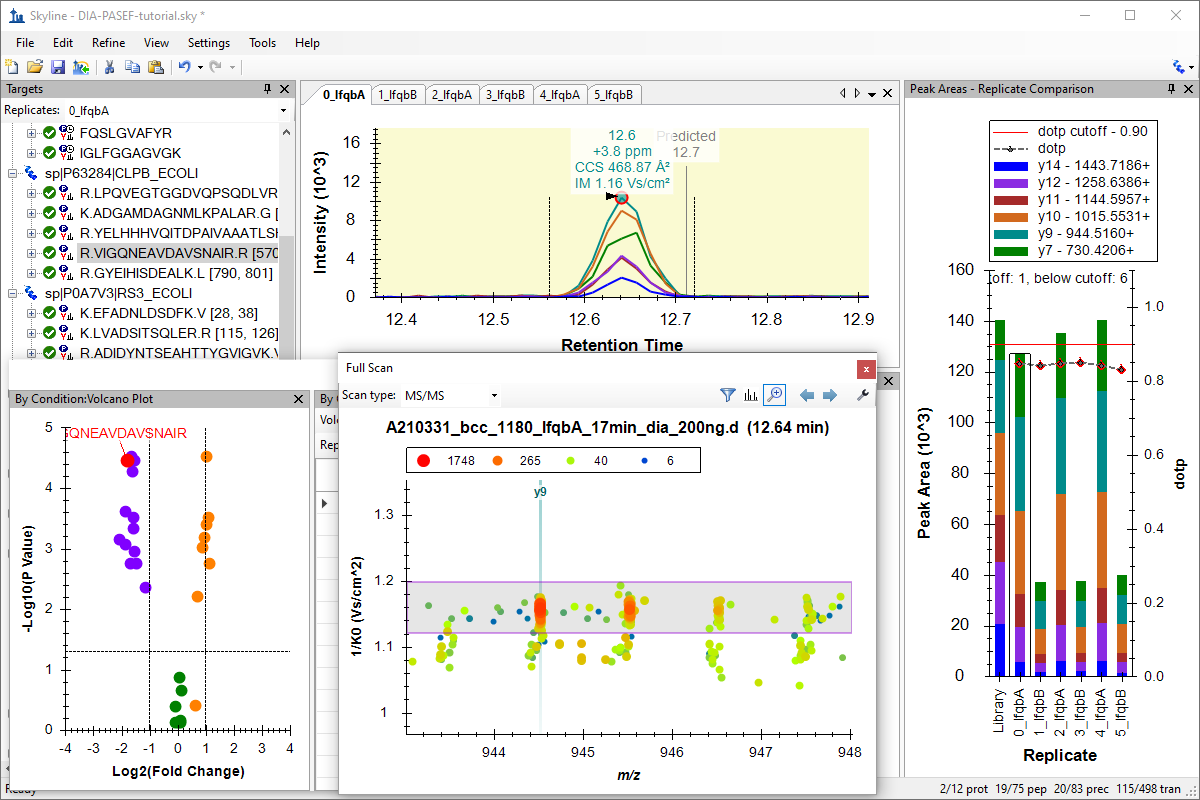
Tutorial: Analysis of diaPASEF Data Work with a data independent acquisition (DIA) parallel accumulation serial fragmentation (PASEF) data, from a Bruker timsTOF instrument, using a 3-organism mix data set created for instruction, based on the Navarro, Nature Biotech 2016 benchmarking paper. Perform a group comparison and make your own assessment of how well the data capture the expected ratios for each organism. (36 pages)
Tutorial: Library-Free DIA/SWATH Run a peptide-spectrum search directly on a data independent acquisition (DIA) dataset, from a SCIEX TripleTOF instrument, using a 3-organism mix data set created for instruction, based on the Navarro, Nature Biotech 2016 benchmarking paper. Use the Run Peptide Search wizard for DIA to run the DIA-Umpire spectrum deconvolution algorithm and then use a traditional spectrum search algorithm to detect peptides in the spectra. (26 pages)
Tutorial: Small Molecule Targets Target small molecule ions with Skyline. Import a small molecule transition list used in a metabolomics experiment and import 14 runs from a Waters Xevo TQS. Start learning how to apply the power of the Skyline interface for small molecule experiments. (9 pages)
Tutorial: Small Molecule Method Development Target stable isotope labeled small molecules from a literature citation, specified as only precursor m/z, product ion m/z, and collision energy values. Learn how many existing Skyline features — created initially for targeted proteomics use — can now be applied to small molecule data. (28 pages)
Tutorial: Small Molecule Multidimension Spectral Library Target lipids in complex ion mobility spectrometry-mass spectrometry (IMS-MS)data using spectral libraries and ion mobility filtering. Explore a spectral library containing m/z, retention time, fragmentation, and ion mobility information, and learn how the CCS values for each molecule are used to increase the selectivity of precursor and fragment extracted ion chromatograms. (23 pages)
Tutorial: Small Molecule Quantification Target small molecules specified as precursor ion chemical formulas and adducts, and product ion m/z values. Import a multi-replicate data set collected using LC-MS/MS on a triple quadulpole, and see how many existing Skyline features created initially for targeted proteomics use can now be applied to small molecule data. (23 pages)
Tutorial: Hi-Res Metabolomics Target small molecules specified as precursor ion chemical formulas and adducts. Import a multi-replicate data set collected on a Q Exactive Orbitrap mass spectrometer for a set of plasma samples, and see how many existing Skyline features created initially for targeted proteomics use can be applied to small molecule data. (18 pages)
Tutorial: Absolute Quantification Use Skyline calibrated quantification to estimate the absolute molecular quantities of peptides in your experiments. (19 pages)
Tutorial: Custom Reports Use custom Live Reports to view, edit and export a wide range of values from your Skyline documents. These reports are perfect for use in Excel or with custom code written in R, Matlab, Java, C++ and other languages for doing deep statistical analysis after processing your instrument output with Skyline. Also learn to use the Skyline Results Grid view to gain access to these values and to add custom annotations while inspecting your data in Skyline. (38 pages)
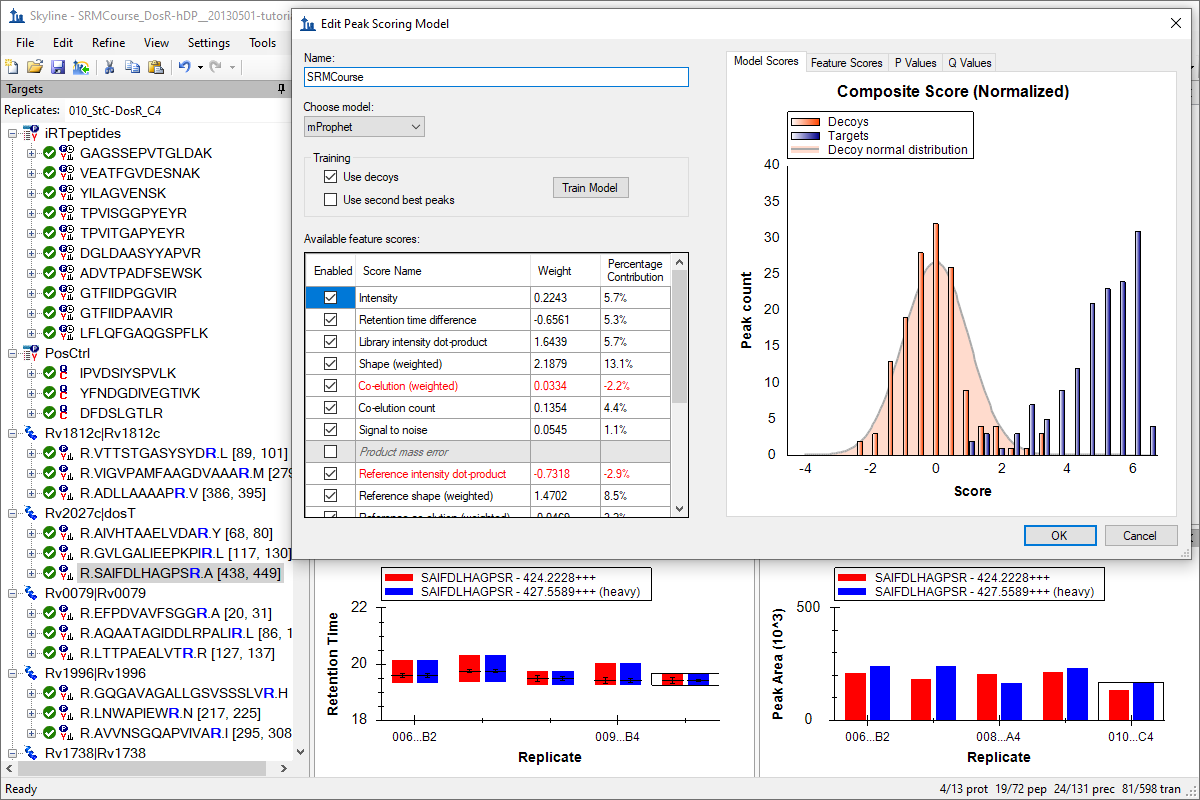
Tutorial: Advanced Peak Picking Models Create advanced models for matching target peptides with chromatogram peaks in Skyline. Skyline supports creating linear combinations of individual peak scores using the mProphet semi-supervised learning algorithm. Learn to generate decoy peptides and transitions, create and assess mProphet scoring models, and apply them to Skyline chromatogram peak picking, both for SRM and DIA/SWATH data. (28 pages)
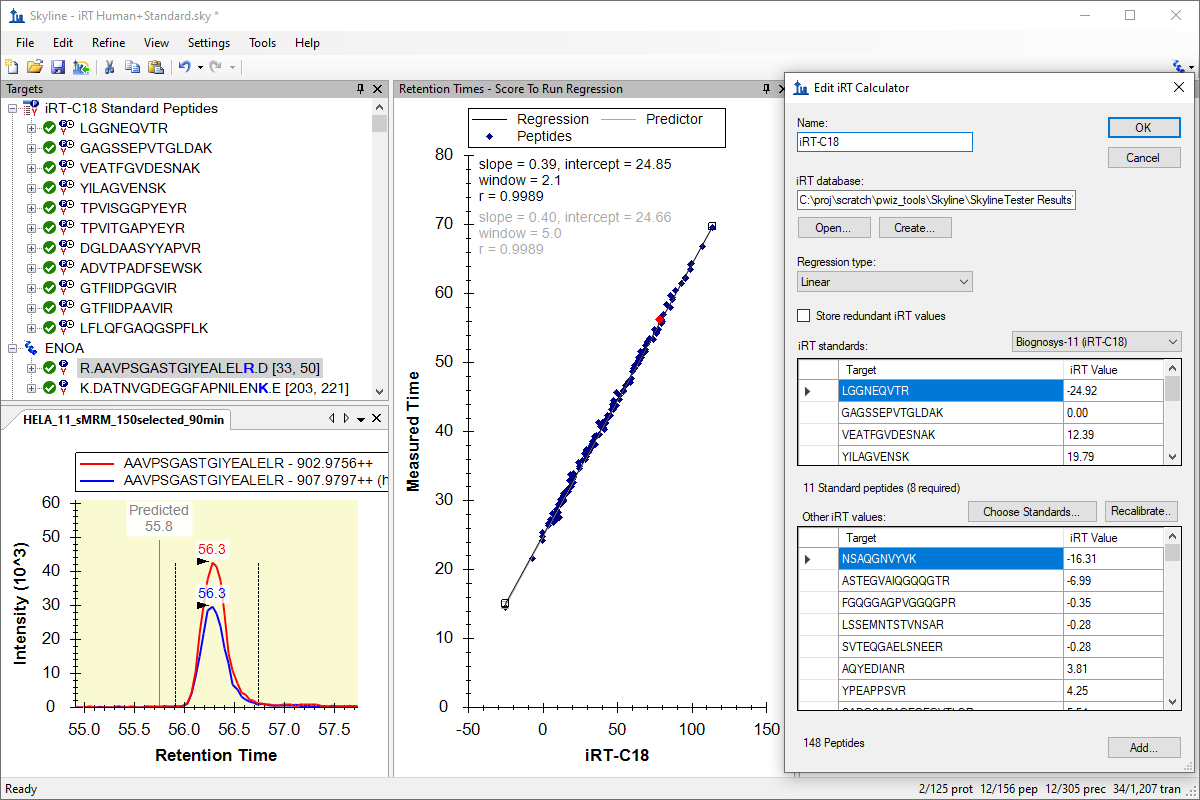
Tutorial: iRT Retention Time Prediction Use iRT in Skyline, a technique for storing calibrated, empirically measured peptide retention times in a library for future use in retention time prediction for scheduled acquisition and peak identity validation. In this tutorial, you will calibrate your own iRT calculator and also learn more about the iRT-C18 calibration proposed by Biognosys for the peptide standards in their iRT-Kit. (31 pages)
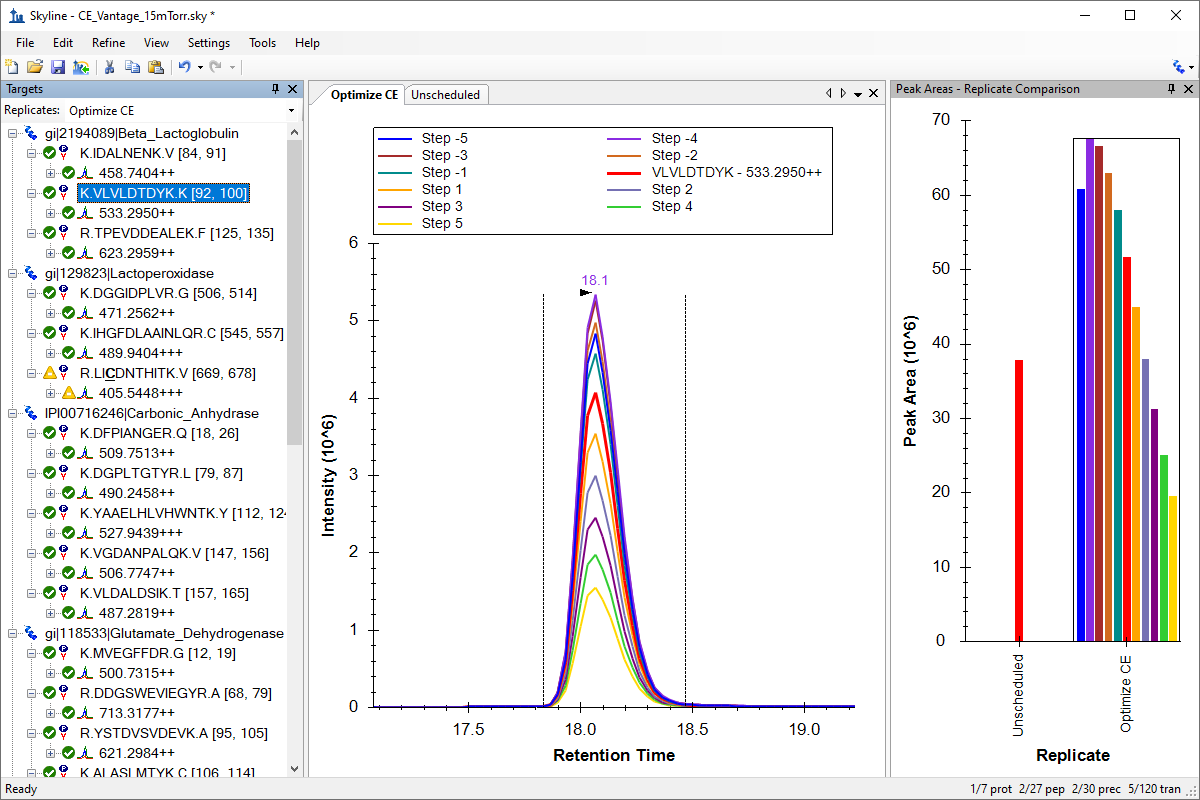
Tutorial: Collision Energy Optimization Use Skyline to optimize collision energy (CE) values. Create scheduled CE optimization transitions lists for a document with 30 peptide precursors. Using supplied RAW files from a Thermo TSQ Vantage, you will recalculate the linear equation used to calculate CE for that instrument. You will also export a transition list with CE values optimized separately for each transition. (12 pages)
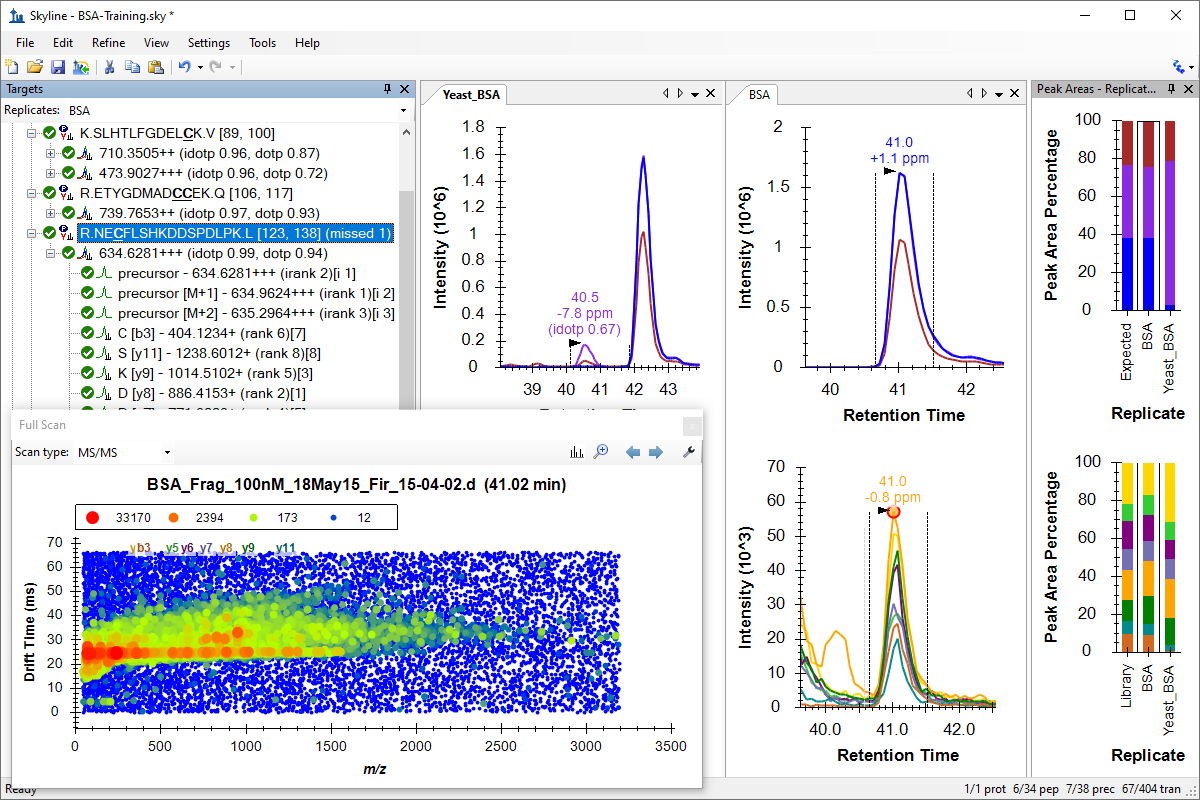
Tutorial: Ion Mobility Spectrum Filtering Use Skyline to work with IMS-TOF data. Train a drift-time predictor for BSA spiked into yeast, and see how ion mobility separation improves the selectivity of chromagram extraction in complex data. Learn how to work with ion mobility data in Skyline and explore the 3D (m/z, IMS, intensity) spectra produced by IMS-enabled mass spectrometers. (26 pages)
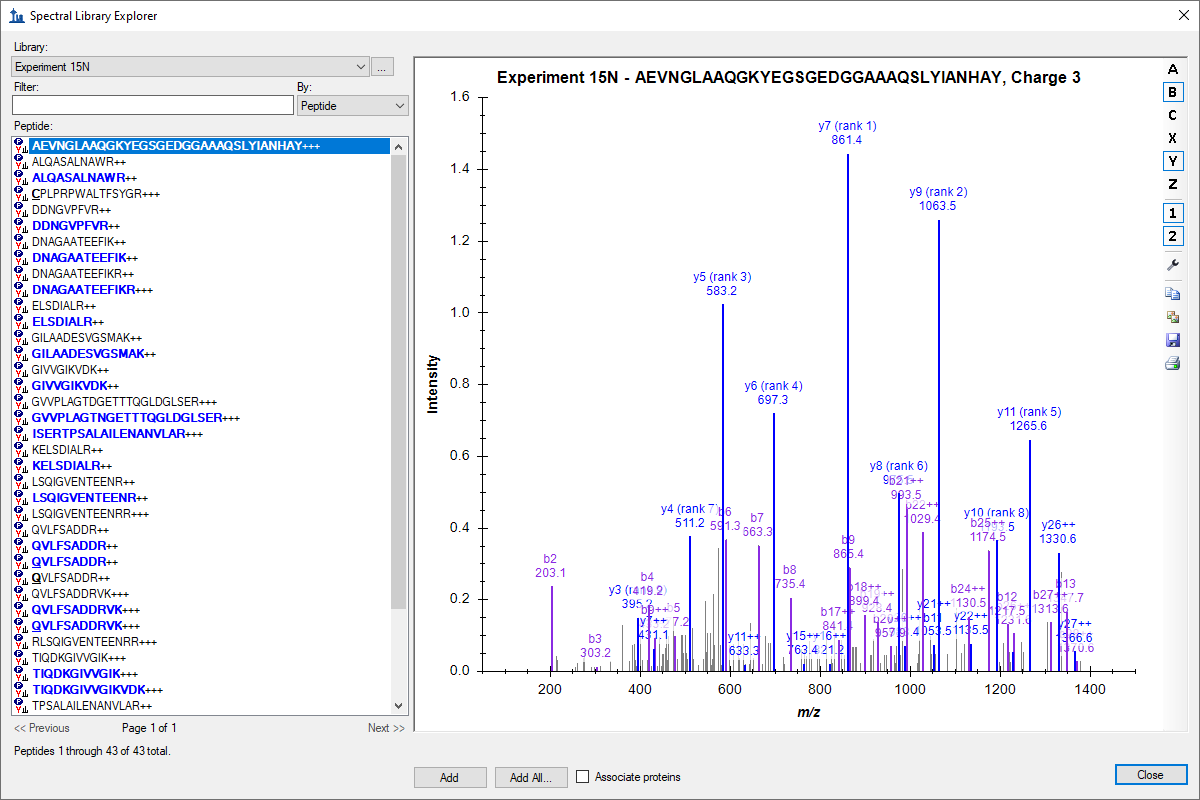
Tutorial: Spectral Library Explorer Use the Skyline Spectral Library Explorer. Learn more about working with isotope labels and product ion neutral losses using MS/MS spectral libraries containing 15N labeled and phosphorylated peptides. Use the Library Explorer to accelerate the transition between shotgun discovery experiments and targeted investigation. (22 pages)
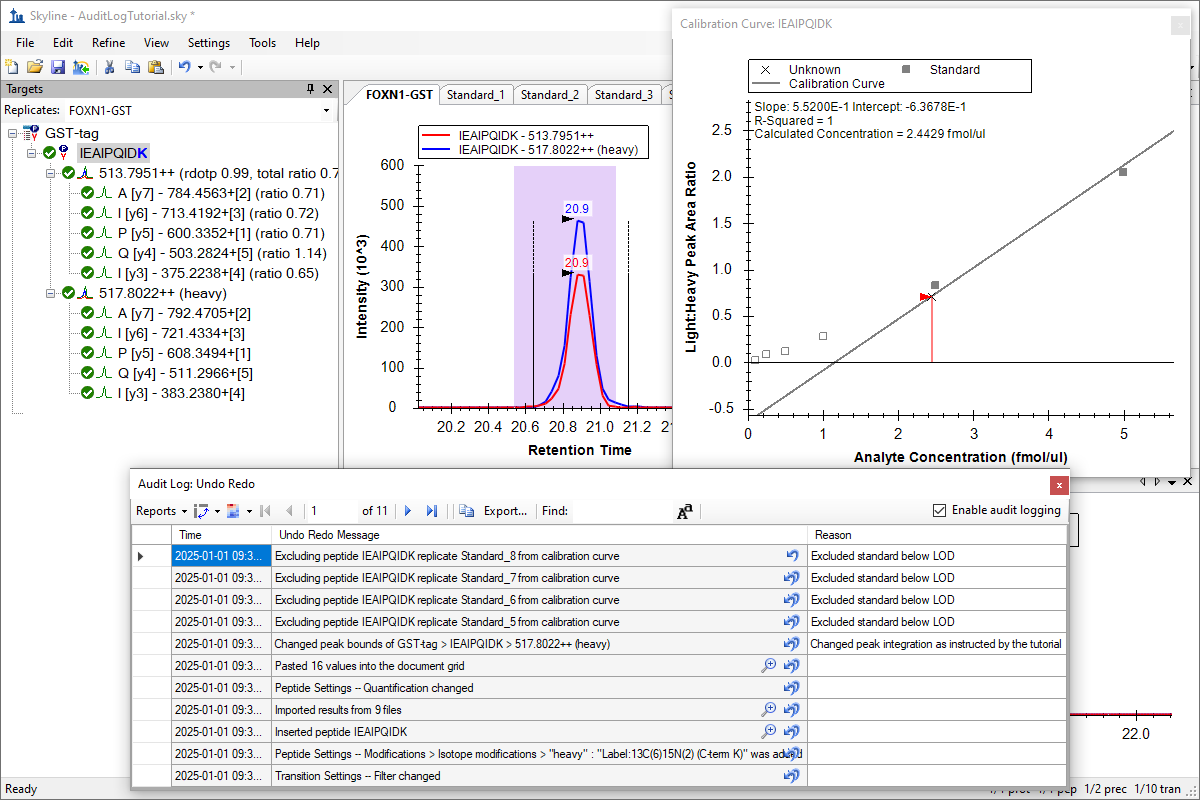
Tutorial: Audit Logging Use audit logging in Skyline. Learn how to produce fully audit logged Skyline documents to help others reproduce your research. Learn how to work with the Audit Log grid view, an extension of the Document Grid to explore logged changes and provide reasons for the changes you make. Finally, you will upload your document to Panorama and see how the audit log is captured for review through a browsable web interface. (23 pages)
|