Skyline Protein Association |
2024-04-20 |
Parsimonious Protein Inference
1. Protein association
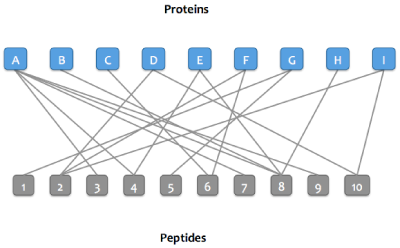
After matching peptides in the Skyline document to proteins in a FASTA or background proteome, a bipartite graph is created with edges between peptides and their matching proteins.
2. Protein grouping
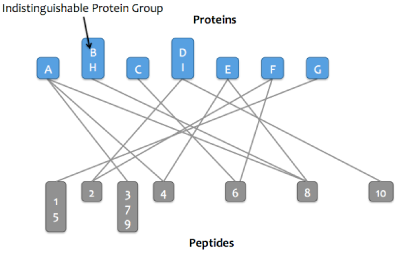
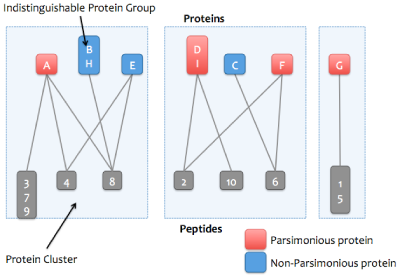
Proteins that match the same set of peptides may optionally be merged into a single node in the graph. These proteins are referred to as an indistinguishable protein group (or just protein group). After this step, the word "protein" may refer to either a protein or a protein group.
3. Sharing peptides between proteins
Many peptides are contained in more than one protein in a FASTA or background proteome. Skyline can optionally assign these peptides to just one protein, or even remove shared peptides entirely. Shared peptides can be:
- Duplicated between proteins
- Assigned to the first protein (that was read in from a FASTA or background proteome)
- Assigned to the protein with the most peptides (in case of ties, all tied proteins will be accepted)
- Removed (peptides must be unique to a protein or protein group)
Before Skyline 22.1, the only two options were "Remove duplicate peptides" and "Remove repeated peptides". Those are still available but have been renamed to "Removed" and "Assigned to first protein".
If neither "Remove subset proteins" nor "Find minimal protein list" are selected, then no more steps are run. Otherwise, step 4 is run next.
4. Protein clustering
To simplify computation and also make it easier for users to understand, the graph is separated into clusters (connected components): these are sets of proteins and peptides that are directly or indirectly connected.
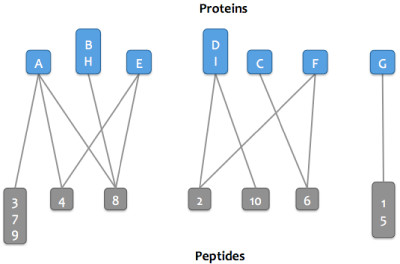
Then either step 4a or step 4b is run depending on which parsimony option is selected.
4a. Finding minimal protein list that explains all peptides
For each cluster, a greedy algorithm is applied that attempts to find the smallest set of proteins that explains all the cluster's peptides. The proteins in the resulting "minimal list" are marked as parsimonious.
Greedy algorithm:
The algorithm works iteratively. For each iteration, the protein that explains the most peptides (that have not previously been explained) is taken out of the cluster and counted as explained. That protein's peptides are also removed from the other proteins that have not yet been considered. When there are no more remaining unexplained peptides, the algorithm is done and the proteins that have not been considered are marked as non-parsimonious. If two or more proteins explain the same number of peptides, then all tied proteins will be accepted into the minimal list. This prevents the parsimony algorithm from arbitrarily excluding proteins due to the order they were enumerated from the FASTA file. The greedy algorithm does not always find the true minimal list, but it's usually pretty close.
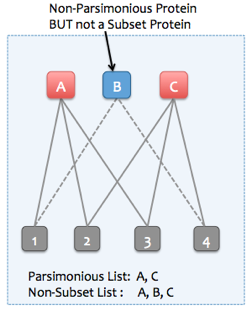
4b. Removing subset proteins
If the "Find minimal protein list that explains all peptides" option is not selected, then Skyline can instead remove only subset proteins. Non-parsimonious, non-subset proteins can be retained (proteins which are not strict subsets of any other protein, but all of their peptides are entirely explained by other parsimonious proteins). To illustrate, consider the following examples:
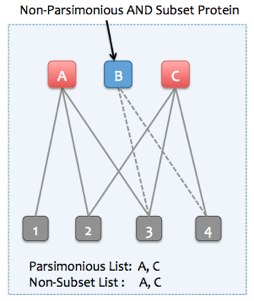
Example of a subset protein (all subset proteins are also non-parsimonious)